[1] "C:/Users/azetner/Documents/quarto-presentations"Best Practices in R
2024-01-31
Best Good Enough Practices in R
Introduction
Seminar Goals
- What to take away from this seminar
- Ideas, resources, methods to improve future work
- What not to take away from this seminar
- Any rush to apply these ideas retroactively to all previous projects
- Inspiration and content from sources listed at the end
Table of Contents
- Project Oriented Workflow
- Readability
- Reproducibility
Project Oriented Workflow
Why?
Work on More Than One Thing at a Time
Team Collaboration
- Easier concurrent work
- Easy distribution
Start and Stop
- Flexible work schedule
- Checkpoint to save progress
Documentation for Continuity
- Easy resumption
- Context and guidelines for collaborators
How?
- Standardized organization of files per project
- Consistent actions
Organization of Project Directories 📂
Organization of Project Directories 📂
- Project Organization:
- Folder per project
- Top-level advertisement
- RStudio/Git/{here} characteristic files
- Path Construction with
here():- Utilize
here()function - here package
- Paths relative to top-level
- Utilize
{here} Package 📂
here() displays top-level folder location
{here} Package 📂
Build a path to a file in a subdirectory and use it
[1] "C:/Users/azetner/Documents/quarto-presentations/presentations/20240131-RBP_images/analysisworkflow.png"[1] "C:/Users/azetner/Documents/quarto-presentations/presentations/20240131-RBP_images"arrow.file <- here("presentations/20240124-BWG_images/arrow_dataset.png")
file.info(arrow.file)["size"] size
C:/Users/azetner/Documents/quarto-presentations/presentations/20240124-BWG_images/arrow_dataset.png 79441Folder Structure 📂
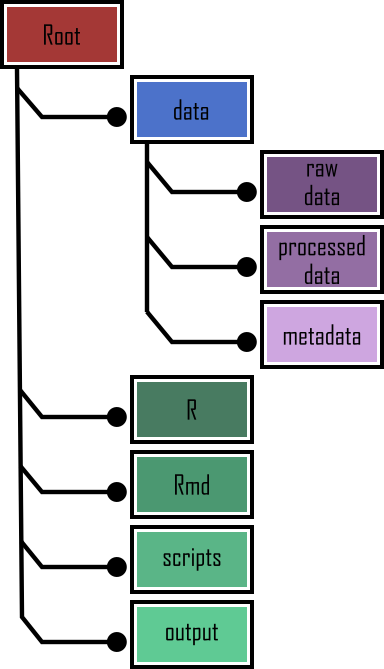
- Folder Structure:
- Data
- Code
- Documentation
- External scripts
- Outputs
- Start project from root
- Console or IDE
here()for paths

RStudio Projects 📂
Maintain project separation
Settings stored in
<NAME>.Rproj.Open Project in RStudio:
- Dedicated R process
- File browser points to Project directory.
- Working directory set to Project.
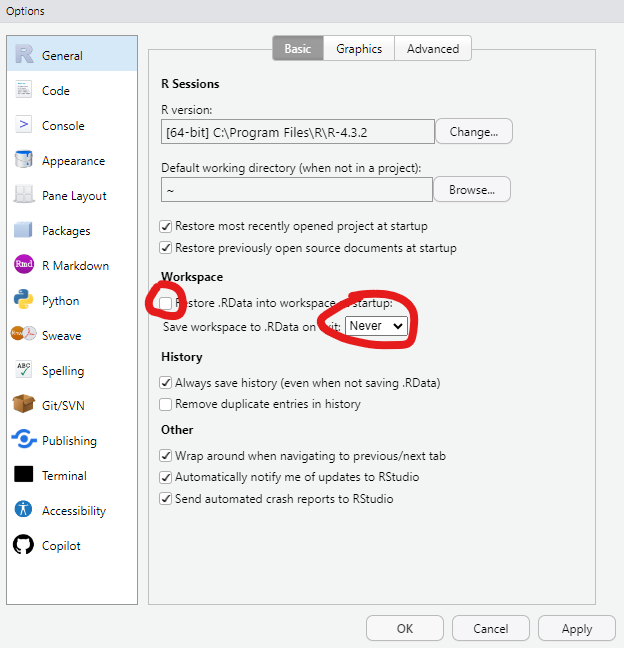
Version: 1.0
RestoreWorkspace: No
SaveWorkspace: No
AlwaysSaveHistory: Default
EnableCodeIndexing: Yes
UseSpacesForTab: Yes
NumSpacesForTab: 2
Encoding: UTF-8
RnwWeave: Sweave
LaTeX: pdfLaTeX
AutoAppendNewline: Yes
StripTrailingWhitespace: Yes
LineEndingConversion: Native
BuildType: Package
PackageUseDevtools: Yes
PackageInstallArgs: --no-multiarch --with-keep.source
PackageRoxygenize: rd,collate,namespaceConsistent Actions 💥
Everything that matters should be achieved through saved code
Save Source not the Workspace / Environment 💥
- Livestock vs. Pets Analogy from Cloud Computing
- Livestock: managed in herds, disposable
- Pets: unique, precious
- Treat R processes like livestock
- Workspace disposability
- Non-reproducible workflows lead to heartache
- Explicitly save important objects
- Checkpoints for long generation time objects
- Design away fear of reproducibility
Use a Blank Slate 💥
R --no-save --no-restore-data
Restart R often 💥
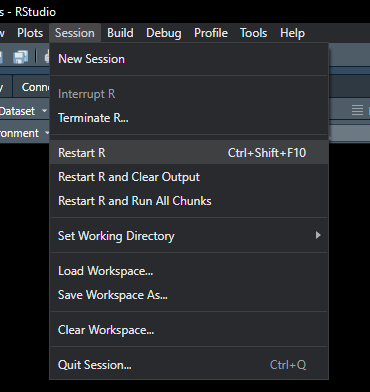
- Restart R to wipe environment
- Save code not workspaces
- Pressure to reinforce correct behaviours
- Ensuring source code recreates important artefacts
Analysis Project Practices ⚡
Use an IDE ⚡
Software Management ⚡
- Research Code and Software:
- Varied forms and sizes
- Includes code processing research data, scripts, and workflows
- Scriptable languages like R, Python, shell, etc
- Standalone programs for specific research tasks
Software Management ⚡
- What can go wrong with research code?
- What does code do?
- Why did we do it this way?
- No longer works
- Accuracy at question
- Software projects all can benefit modular code:
- Readable
- Reusable
- Testable
Software Management ⚡

Comment brief explanations
Functions first
- Clear inputs and outputs
- Meaningful names
- One main task
Ruthlessly eliminate duplication
- Functions
- Data Structures
- Effort
Software Management ⚡

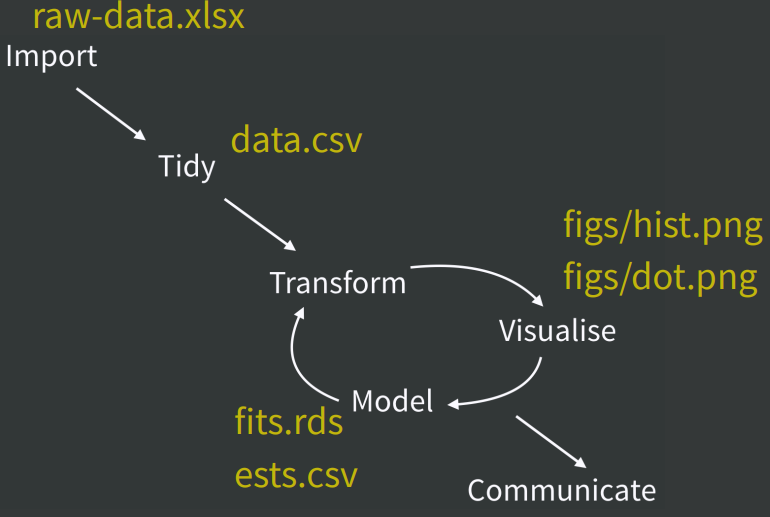
Data Management 💽
- Why Data Management?
- Data loss / corruption
- Confusion about provenance
- Version
Data Management 💽
- Data Management
- Save the raw data
- Ensure raw data is backed up
- Create analysis-friendly data
- Create the data you wish to see in the world
- Self explanatory naming
- Open formats
- Machine readability
- Export cleaned data that you wish you’d received
- Create the data you wish to see in the world
- Record all the steps used to process data
Data Management 💽

- Each variable must have its own column
- Each observation must have its own row
- Each value must have its own cell
- Consistency
- Vectorization
Data Management 💽

Readability 📑
Naming Conventions 📑
- File names should be:
- Machine readable
- Human readable
- Optional: Consistent
- Optional: Play well with default ordering
Machine Readable 📑
- Regex/Globbing Friendly
- Avoid spaces, punctuation, and accented characters
- Maintain case sensitivity
- Easy Computation
- Use intentional delimiters for straightforward computational processes
- Deliberate delimiter use enhances computational efficiency
- Dashes for spaces between words
- Underscores for chunks
20220120_patient-exposure_control.csv
20220120_patient-exposure_treatment.csv
20220215_patient-exposure_control.csv
20220215_patient-exposure_treatment.csv
20220215_patient-info_control.csv
20220310_patient-info_control.csv
20220520_patient-info_treatment.csv
20220805_patient-info_control.csv
20230120_patient-info_treatment.csv
20230215_patient-info_treatment.csv
20230310_patient-exposure_control.csv
20230310_patient-exposure_treatment.csv
20230405_patient-exposure_control.csv
20230405_patient-exposure_treatment.csv
20230405_patient-info_control.csv
20230615_patient-info_treatment.csv
20230710_patient-info_control.csv
20230710_patient-info_treatment.csv
20230805_patient-info_treatment.csv❯ ls -1 2022*
20220120_patient-exposure_control.csv
20220120_patient-exposure_treatment.csv
20220215_patient-exposure_control.csv
20220215_patient-exposure_treatment.csv
20220215_patient-info_control.csv
20220310_patient-info_control.csv
20220520_patient-info_treatment.csv
20220805_patient-info_control.csv❯ ls -1 *info*
20220215_patient-info_control.csv
20220310_patient-info_control.csv
20220520_patient-info_treatment.csv
20220805_patient-info_control.csv
20230120_patient-info_treatment.csv
20230215_patient-info_treatment.csv
20230405_patient-info_control.csv
20230615_patient-info_treatment.csv
20230710_patient-info_control.csv
20230710_patient-info_treatment.csv
20230805_patient-info_treatment.csvHuman Readable 📑
- Informative File Names:
- Include content information in file names
- Anticipate usage context
- Slug:
- User-friendly and descriptive filenames
20230710_*patient-info_control*.csv

filedir <- here("presentations/20240131-RBP_images/fakedat/")
flist <- list.files(filedir, pattern = "info")
stringr::str_split_fixed(flist, "[_\\.]", 4) [,1] [,2] [,3] [,4]
[1,] "20220215" "patient-info" "control" "csv"
[2,] "20220310" "patient-info" "control" "csv"
[3,] "20220520" "patient-info" "treatment" "csv"
[4,] "20220805" "patient-info" "control" "csv"
[5,] "20230120" "patient-info" "treatment" "csv"
[6,] "20230215" "patient-info" "treatment" "csv"
[7,] "20230405" "patient-info" "control" "csv"
[8,] "20230615" "patient-info" "treatment" "csv"
[9,] "20230710" "patient-info" "control" "csv"
[10,] "20230710" "patient-info" "treatment" "csv"
[11,] "20230805" "patient-info" "treatment" "csv"Easy Sorting 📑
- Numeric Inclusion:
- Often for code
- Include a numeric element for effective sorting
- Left-pad with zeros for consistent width and visual sorting.
- eg
01_import.R
- Dates:
- Utilize the ISO 8601 standard for date formatting: YYYYMMDD
- Ensures chronological sorting in file names by default
- eg
20220820_wedding-photos.zip
Naming Conventions 📑
- Avoid:
- Internal sequential numbers:
result1.csv,result2.csv - Manuscript locations:
fig_3_a.png
- Internal sequential numbers:
Writing Meaningful Comments 💬
Programs must be written for people to read, and only incidentally for machines to execute.
Meaningful Comments 💬
Rule 1: Comments should not duplicate the code

Meaningful Comments 💬
Rule 2: Good comments do not excuse unclear code.

Meaningful Comments 💬
Rule 3: If you can’t write a clear comment, there may be a problem with the code.
“Debugging is twice as hard as writing the code in the first place. Therefore, if you write the code as cleverly as possible, you are, by definition, not smart enough to debug it.”
Roxygen2 Comments 💬

Roxygen2 Comments 💬
- In-line documentation
- Rich, dynamic
.Rdfile generation - Standard markdown
% Generated by roxygen2: do not edit by hand
% Please edit documentation in R/add.R
\name{add}
\alias{add}
\title{Add together two numbers}
\usage{
add(x, y)
}
\arguments{
\item{x}{A number.}
\item{y}{A number.}
}
\value{
A numeric vector.
}
\description{
Add together two numbers
}
\examples{
add(1, 1)
add(10, 1)
}Roxygen2 Comments 💬
Reproducibility 🐣
Modular and Reusable Code 🐣
- Start programs/functions with explanatory comments.
- Use meaningful names for functions and variables.
- Explicitly state dependencies and requirements.
- Avoid commenting/uncommenting code sections for program control.
- Include a simple example or test data set for clarity.
Make an R Package 🐣
- Why make an R package?
- Code Organization
- Consistent documentation
- Code Distribution
- Dependency handling
Make an R Package 🐣
- Package directory structure
- Scripts in
R - Documentation in
man - README.md
- DESCRIPTION
- NAMESPACE
- Scripts in
- {Roxygen2} for documentation
- {devtools} for process
.
├── DESCRIPTION
├── NAMESPACE
├── R
│ ├── cat_function.R
│ ├── cat_images.R
│ └── cats-package.R
├── README.md
└── man
├── add_cat.Rd
├── cat_function.Rd
├── cats.Rd
├── get_cat.Rd
└── here_kitty.RdMake an R Package 🐣
Make an R Package 🐣
- DESCRIPTION file
- Dependencies
- Package metadata
- Contact information
Package: cats
Title: Cats
Version: 0.1
Author: Hilary Parker <hilary@etsy.com> [aut, cre]
Maintainer: Hilary Parker <hilary@etsy.com>
Authors@R: c( person("Hilary", "Parker", email = "hilary@etsy.com", role =
c("aut", "cre")))
Description: Mew.
Depends:
R (>= 3.0.2)
Imports:
httr,
ggplot2,
jpeg
License: MIT
LazyData: true
Suggests:
testthatMake an R Package 🐣
- NAMESPACE file
- Functions from your package
- Imported functions from others
# Generated by roxygen2 (4.0.1.99): do not edit by hand
export(add_cat)
export(cat_function)
export(get_cat)
export(here_kitty)
import(ggplot2)
import(httr)
import(jpeg)Version Control 🧭
Why version control in data analysis projects?
- File Versioning
- Figure Recreation
- Code Modification Impact
- Directory Copying Fear
- Shared File Row Duplication
- Lost Data Files
- Analysis Choice
- Manuscript Collaboration Merge
- Accidental Deletion
- Project Status Recall
- Experiment Mistake Identification
- Directory Pollution
- Selective Changes
- Ad Infinitum
Version Control 🧭
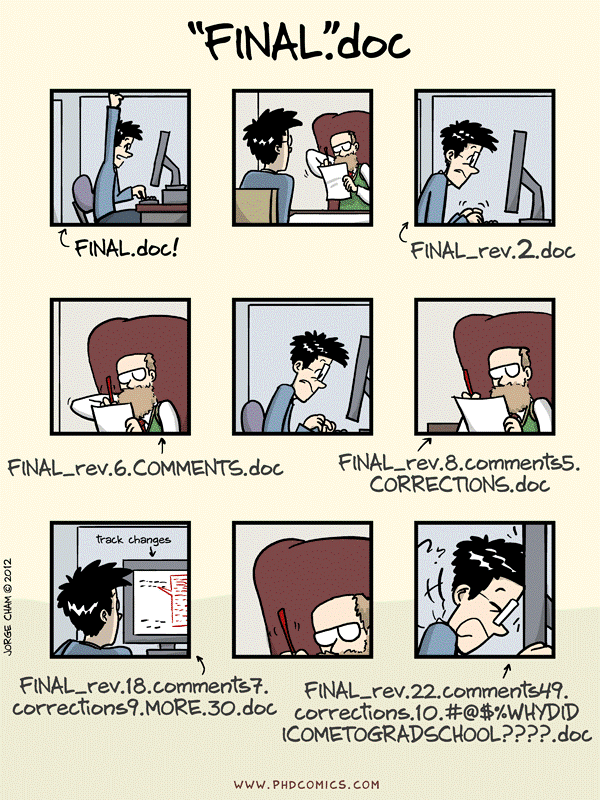
Version Control 🧭
- What to Save:
- Human-created content.
- Plain text files.
- Avoid binary files.
Version Control: Manual 🧭
CHANGELOG.txt
## 2016-04-08
* Switched to cubic interpolation as default.
* Moved question about family's TB history to end of questionnaire.
## 2016-04-06
* Added option for cubic interpolation.
* Removed question about staph exposure (can be inferred from blood test results).Version Control: Manual 🧭
Backup entire project folder
.
|-- project_name
| -- current
| -- ...project content as described earlier...
| -- 2016-03-01
| -- ...content of 'current' on Mar 1, 2016
| -- 2016-02-19
| -- ...content of 'current' on Feb 19, 2016… please don’t do this
Version Control: VCS 🧭
- Version Control System
- Efficient Backup Storage
- Automated Timestamps
- Automated Changelog and Accuracy
- Conflict Resolution and Merging
- Examples: Git, SVN, Bitbucket, etc
Version Control: VCS 🧭


Version Control: VCS 🧭
Managing Project Dependencies 📦
The only way to be certain that code written by someone else will run on your machine and will produce the same results is to replicate their environment
Managing Project Dependencies 📦
- What is the environment?
- The version of the software used
- R 3.6.1 vs R 4.3.2 - The versions of the packages/extensions used
- {dplyr} 1.0.10 vs 1.1.12 - External software dependencies for packages
- pandoc
- The version of the software used
Resources
http://tinyurl.com/COGseminar

Seminar Content
Moodle Cources
Readability
Packages
Project Dependencies
Useful Links
